
- Sanger sequence analysis how to#
- Sanger sequence analysis software#
- Sanger sequence analysis password#
- Sanger sequence analysis download#
Sanger sequence analysis software#
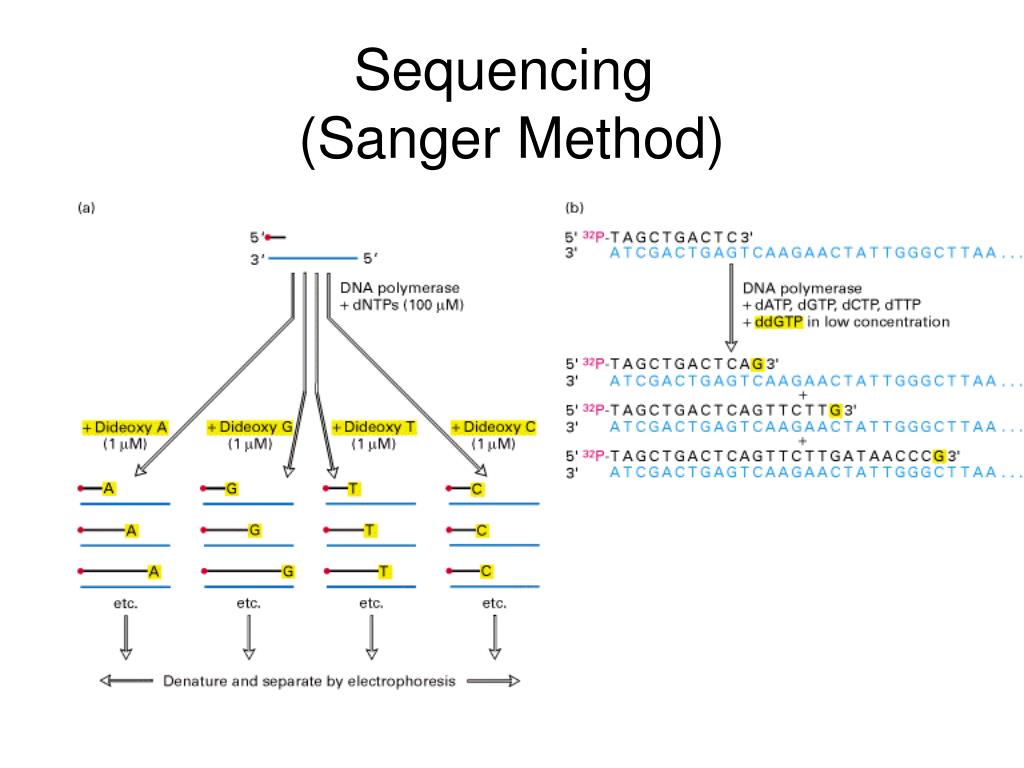
There are many software tools for the analysis of Sanger raw files. For these and other cases, Sanger sequencing remains a grateful option, as it is relatively inexpensive and delivers results relatively quickly. Sanger Sequencing is performed on an Applied Biosystems 3730 Genetic Analyzer, a 48 capillary electrophoresis instrument for DNA sequencing or DNA fragment. Frequently Asked QuestionsĬommon questions regarding this topic have been collected on a dedicated FAQ page. Finally, Sanger sequencing is frequently used to verify mutations identified by NGS. Developed by two-time Nobel laureate Frederick Sanger and his colleagues in 1977, it enabled an international collaboration of scientists to deliver the first human genome sequence.
Sanger sequence analysis password#
Login as admin with password password to access everything. Sanger sequencing is a method for determining the nucleotide sequence of DNA molecules. It will launch a flavored Galaxy instance available on This instance will contain all the tools and workflows to follow the tutorials in this topic.
Sanger sequence analysis download#
NOTE: Use the -d flag at the end of the command if you want to automatically download all the data-libraries into the container.
Sanger sequence analysis how to#
If you need help with iLab the Seqlab staff is happy to help you get started! Below are instructions on how to register to iLab and make a work request for Sanger and fragment analysis services.Docker run -p 8080:80 quay.io/galaxy/sequence-analysis-training The old shared Seqlab email address is no longer in use - you can reach Seqlab staff via their personal email addresses. We are also using iLab as the primary communication channel regarding possible breaks in sample processing due to instrument maintenance, staff holidays etc. In iLab you will find the sample submission form along with information about sample processing and up to date pricing for available services. SeqLab uses HiLife iLab online system for managing orders and invoices. It can be used for DNA sequencing, genotyping and fragment. Maximum read length (QV>20) for high quality sequencing product is around 600bp. The BAS Lab has an automated 3130XL Genetic analyzer which has a 16 capillary 50 cm array. As a starting material we use purified PCR products mixed with sequencing primer or ready made sequencing/fragment analysis reactions. When troubleshooting sequencing data, follow the workflow below to try to identify the cause of your problem. Sequencing system used is Applied Biosystems ABI3730XL DNA Analyzer. Sanger Sequencing Troubleshooting Guide (GNGFM00346) v1.1. In addition to DNA sequencing, small to medium scale fragment analysis service is also available.


 0 kommentar(er)
0 kommentar(er)
